Which Dna Strand Is Synthesized in a Continuous Fashion
Watson and Crick's discovery of DNA structure in 1953 revealed a possible mechanism for Dna replication. So why didn't Meselson and Stahl finally explain this mechanism until 1958?
This structure has novel features which are of considerable biological interest . . . It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
—Watson & Crick (1953)
Perhaps the most significant aspect of Watson and Crick'southward discovery of Deoxyribonucleic acid construction was not that it provided scientists with a iii-dimensional model of this molecule, but rather that this structure seemed to reveal the way in which DNA was replicated. As noted in their 1953 newspaper, Watson and Crick strongly suspected that the specific base pairings within the Deoxyribonucleic acid double helix existed in order to ensure a controlled organisation of DNA replication. However, it took several years of subsequent study, including a archetype 1958 experiment by American geneticists Matthew Meselson and Franklin Stahl, before the exact relationship between DNA structure and replication was understood.
Three Proposed Models for DNA Replication
Replication is the process past which a cell copies its DNA prior to division. In humans, for example, each parent cell must copy its unabridged half-dozen billion base pairs of Dna before undergoing mitosis. The molecular details of Dna replication are described elsewhere, and they were not known until some fourth dimension after Watson and Crick'southward discovery. In fact, before such details could be determined, scientists were faced with a more than fundamental inquiry concern. Specifically, they wanted to know the overall nature of the process by which DNA replication occurs.
Defining the Models
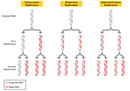
Equally previously mentioned, Watson and Crick themselves had specific ideas most DNA replication, and these ideas were based on the structure of the DNA molecule. In detail, the duo hypothesized that replication occurs in a "semiconservative" manner. According to the semiconservative replication model, which is illustrated in Figure 1, the two original Dna strands (i.e., the two complementary halves of the double helix) separate during replication; each strand and so serves as a template for a new DNA strand, which ways that each newly synthesized double helix is a combination of one old (or original) and one new DNA strand. Conceptually, semiconservative replication made sense in light of the double helix structural model of Deoxyribonucleic acid, in particular its complementary nature and the fact that adenine always pairs with thymine and cytosine always pairs with guanine. Looking at this model, it is like shooting fish in a barrel to imagine that during replication, each strand serves every bit a template for the synthesis of a new strand, with complementary bases existence added in the order indicated.
Semiconservative replication was non the only model of Dna replication proposed during the mid-1950s, withal. In fact, two other prominent hypotheses were put likewise along: conservative replication and dispersive replication. According to the bourgeois replication model, the entire original Dna double helix serves equally a template for a new double helix, such that each circular of cell division produces one daughter cell with a completely new DNA double helix and some other daughter prison cell with a completely intact old (or original) DNA double helix. On the other hand, in the dispersive replication model, the original Dna double helix breaks apart into fragments, and each fragment then serves every bit a template for a new DNA fragment. As a upshot, every prison cell division produces two cells with varying amounts of onetime and new Deoxyribonucleic acid (Effigy 1).
Making Predictions Based on the Models
When these 3 models were first proposed, scientists had few clues about what might be occurring at the molecular level during DNA replication. Fortunately, the models yielded different predictions nearly the distribution of erstwhile versus new DNA in newly divided cells, no matter what the underlying molecular mechanisms. These predictions were as follows:
- Co-ordinate to the semiconservative model, after i round of replication, every new Deoxyribonucleic acid double helix would be a hybrid that consisted of 1 strand of one-time Deoxyribonucleic acid leap to one strand of newly synthesized Dna. And so, during the second round of replication, the hybrids would divide, and each strand would pair with a newly synthesized strand. Afterward, just half of the new DNA double helices would be hybrids; the other half would be completely new. Every subsequent circular of replication therefore would result in fewer hybrids and more than completely new double helices.
- Co-ordinate to the conservative model, afterwards one round of replication, half of the new DNA double helices would be composed of completely erstwhile, or original, DNA, and the other half would exist completely new. Then, during the 2d round of replication, each double helix would be copied in its entirety. Later on, one-quarter of the double helices would be completely quondam, and iii-quarters would exist completely new. Thus, each subsequent round of replication would outcome in a greater proportion of completely new DNA double helices, while the number of completely original Dna double helices would remain constant.
- Co-ordinate to the dispersive model, every circular of replication would issue in hybrids, or DNA double helices that are part original DNA and office new Dna. Each subsequent circular of replication would so produce double helices with greater amounts of new DNA.
Meselson and Stahl'due south Elegant Experiment
Matthew Meselson and Franklin Stahl were well acquainted with these iii predictions, and they reasoned that if there were a style to distinguish old versus new DNA, it should be possible to exam each prediction. Enlightened of previous studies that had relied on isotope labels as a style to differentiate between parental and progeny molecules, the scientists decided to see whether the same technique could be used to differentiate between parental and progeny DNA. If it could, Meselson and Stahl were hopeful that they would be able to decide which prediction and replication model was correct.
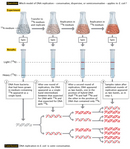
The duo thus began their experiment by choosing two isotopes of nitrogen—the mutual and lighter 14North, and the rare and heavier 15North (and so-chosen "heavy" nitrogen)—every bit their labels and a technique known as cesium chloride (CsCl) equilibrium density gradient centrifugation as their sedimentation method. Meselson and Stahl opted for nitrogen because it is an essential chemical component of DNA; therefore, every time a jail cell divides and its Dna replicates, it incorporates new Due north atoms into the DNA of either one or both of its two daughter cells, depending on which model was correct. "If several unlike density species of Dna are nowadays," they predicted, "each volition form a band at the position where the density of the CsCl solution is equal to the buoyant density of that species. In this fashion, DNA labeled with heavy nitrogen (xvN) may be resolved from unlabeled Dna" (Meselson & Stahl, 1958).
The scientists and so continued their experiment by growing a culture of East. coli bacteria in a medium that had the heavier fifteenNorth (in the form of 15North-labeled ammonium chloride) as its only source of nitrogen. In fact, they did this for 14 bacterial generations, which was long enough to create a population of bacterial cells that independent only the heavier isotope (all the original fourteenNorthward-containing cells had died by then). Next, they inverse the medium to one containing simply 14North-labeled ammonium salts as the sole nitrogen source. So, from that point onward, every new strand of DNA would be built with 14N rather than 15Due north.
Simply prior to the addition of 14N and periodically thereafter, as the bacterial cells grew and replicated, Meselson and Stahl sampled Dna for use in equilibrium density gradient centrifugation to determine how much fifteenDue north (from the original or erstwhile DNA) versus 14N (from the new DNA) was present. For the centrifugation procedure, they mixed the Deoxyribonucleic acid samples with a solution of cesium chloride and then centrifuged the samples for plenty time to allow the heavier fifteenDue north and lighter xivN DNA to drift to different positions in the centrifuge tube.
By way of centrifugation, the scientists found that DNA composed entirely of fifteenN-labeled DNA (i.e., DNA nerveless only prior to changing the civilization from one containing simply 15N to one containing only fourteenN) formed a unmarried distinct band, because both of its strands were made entirely in the "heavy" nitrogen medium. Following a unmarried round of replication, the DNA over again formed a unmarried distinct band, but the band was located in a different position along the centrifugation gradient. Specifically, information technology was found midway between where all the 15Northward and all the 14Due north DNA would have migrated—in other words, halfway between "heavy" and "lite" (Effigy 2). Based on these findings, the scientists were immediately able to exclude the conservative model of replication as a possibility. Subsequently all, if Dna replicated conservatively, in that location should have been two distinct bands after a unmarried round of replication; one-half of the new DNA would have migrated to the aforementioned position as it did earlier the civilisation was transferred to the 14Northward-containing medium (i.e., to the "heavy" position), and only the other half would have migrated to the new position (i.e., to the "light" position). That left the scientists with just ii options: either Dna replicated semiconservatively, as Watson and Crick had predicted, or it replicated dispersively.
To differentiate between the two, Meselson and Stahl had to let the cells separate again and then sample the Dna after a second circular of replication. Subsequently that second round of replication, the scientists institute that the Dna separated into two singled-out bands: i in a position where Deoxyribonucleic acid containing only 14N would be expected to migrate, and the other in a position where hybrid DNA (containing half fourteenDue north and half fifteenN) would exist expected to migrate. The scientists connected to observe the same ii bands after several subsequent rounds of replication. These results were consistent with the semiconservative model of replication and the reality that, when DNA replicated, each new double helix was built with 1 erstwhile strand and one new strand. If the dispersive model were the correct model, the scientists would accept continued to discover merely a unmarried band afterwards every round of replication.
Direct or Circular?
Following publication of Meselson and Stahl's results, many scientists confirmed that semiconservative replication was the rule, not just in E. coli, but in every other species studied as well. To date, no one has found any bear witness for either conservative or dispersive DNA replication. Scientists have institute, however, that semiconservative replication tin can occur in different means—for example, it may continue in either a circular or a linear fashion, depending on chromosome shape.
In fact, in the early on 1960s, English molecular biologist John Cairns performed another remarkably elegant experiment to demonstrate that E. coli and other bacteria with circular chromosomes undergo what he termed "theta replication," because the construction generated resembles the Greek alphabetic character theta (Θ). Specifically, Cairns grew E. coli leaner in the presence of radioactive nucleotides such that, later replication, each new Deoxyribonucleic acid molecule had one radioactive (hot) strand and one nonradioactive strand. He then isolated the newly replicated Dna and used it to produce an electron micrograph epitome of the Θ-shaped replication process (Figure 3; Cairns, 1961).
Only how does theta replication work? It turns out that this process results from the original double-stranded Dna unwinding at a single spot on the chromosome known as the replication origin. Equally the double helix unwinds, it creates a loop known as the replication chimera, with each newly separated single strand serving as a template for Deoxyribonucleic acid synthesis. Replication occurs as the double helix unwinds.
Eukaryotes undergo linear, not circular, replication. As with theta replication, as the double helix unwinds, each newly separated single strand serves as a template for DNA synthesis. All the same, unlike bacterial replication, because eukaryotic cells deport vastly more than DNA than bacteria do (for example, the common house [and laboratory] mouse Mus musculus has almost 3 billion base pairs of Dna, compared to a bacterial cell's i to four million base pairs), eukaryotic chromosomes have multiple replication origins, with multiple replication bubbling forming. For example, K. musculus has as many as 25,000 replication origins, whereas the smaller-genomed fruit fly ( Drosophila melanogaster ), with its approximately 120 million base of operations pairs of Deoxyribonucleic acid, has only about 3,500 replication origins.
Thus, the discovery of the structure of DNA in 1953 was only the beginning. When Watson and Crick postulated that form predicts function, they provided the scientific customs with a challenge to determine exactly how DNA functioned in the cell, including how this molecule was replicated. The piece of work of Meselson and Stahl demonstrates how elegant experiments can distinguish betwixt unlike hypotheses. Understanding that replication occurs semiconservatively was just the beginning to understanding the key enzymatic events responsible for the concrete copying of the genome.
References and Recommended Reading
Cairns, J. The bacterial chromosome and its manner of replication as seen past autoradiography. Periodical of Molecular Biology half-dozen, 208–213 (1961)
Meselson, M., & Stahl, F. The replication of Deoxyribonucleic acid in Escherichia coli. Proceedings of the National University of Sciences 44, 671–682 (1958)
Watson, J. D., & Crick, F. H. C. A construction for deoxyribose nucleic acid. Nature 171, 737–738 (1953) (link to article).

This content is currently under construction.
Explore This Subject
Applications in Biotechnology
Transcription & Translation
Discovery of Genetic Fabric
Topic rooms within Nucleic Acid Structure and Role
Shut
No topic rooms are there.


- | Lead Editor: Bob Moss
0 Response to "Which Dna Strand Is Synthesized in a Continuous Fashion"
Post a Comment